A FULLY PHASED GENOME ASSEMBLY FOR Mytilus edulis UNVEILS A HIGH DEGREE OF PRESENCE-ABSENCE VARIANCE BETWEEN MUSSEL POPULATIONS
Mussels belonging to the Mytilus species complex are cultivated worldwide, and Prince Edward Island produces 80% of the mussels sold in North America. Bivalve genomes are complex and contain many paralogous regions that can confound the separation of all types of variants in a genome. In addition, the mussel genome is highly repetitive and heterozygous.
To overcome the challenges imposed by these characteristics, we used a hybrid assembly approach combining PacBio CLR sequencing, Dovetail Omni-seq scaffolding, PacBio Hi-Fi sequencing and PacBio IsoSeq. We present a fully-phased chromosome level assembly of the mussel’s genome that enabled the genome-wide evaluation of presence-absence variance in Mytilus edulis.
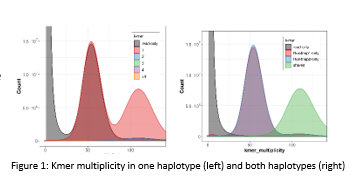
Length and contiguity metrics were: number of scaffolds = 347; N50 = 105 Mb, NG50 = 150 Mb, Total Length = 1.58 Gb. Quality Values and completeness generated using Merqury indicated that each haplotype individually only contains ~65% of the kmers present in the raw HiFi reads, but combined both haplotypes contain ~99% of the kmers present in the raw reads. Our haplotype collapsed assembly available on GenBank only contains 75% of the kmer present in the raw reads. That indicated that up to 25% of the polymorphism variation may be lost in a collapsed assembly.
BUSCO analysis shows that the primary assembly contains 98% of the eukaryotic conserved orthologs in odb10. PAV analysis indicates that up to 13% of shotgun reads from different individuals do not map to the reference genome. Ab-Initio annotation produced 41,319 gene models. Contigs from assembled unmapped reads contained 65,996 putative transcripts while the reference transcriptome generated with Isoseq clustering identified 216,434 putative transcripts, indicating that up to 30% of the mussel pan-transcriptome could be disposable.
In conclusion, we presented a road map to producing high-quality chromosome level phased assemblies for mussels. We also demonstrated the value of haplotype resolved assemblies for genomic analysis in blue mussels and showed evidence of significant PAV among different mussel individuals.